
METU HS Ankara 2015 Team
VARROA CULA


Parts
We successfully characterized our devices and composite parts.

Table 1. Overview
Cinnamyl Alcohol Dehydroganase (CAD)
Length: 1074 bp
Function:
In short: Involved in lignin biosynthesis. Catalyzes the final step specific for the production of lignin monomers. Catalyzes the NADPH-dependent reduction of coniferaldehyde, 5-hydroxyconiferaldehyde, sinapaldehyde, 4-coumaraldehyde and caffeyl aldehyde to their respective alcohols.
Detailed: Cinnamyl alcohol dehydrogenase (CAD, EC 1.1.1.195) is an NADP(H) specific oxidoreductase catalysing the reversible conversion of cinnamyl aldehydes to the corresponding alcohols or monolignols (Mansel et al., 1974; Wyrambik and Griesebach, 1975; Boudet et al., 1995). Polymerization of the three monolignols: coumaryl, sinapyl and coniferyl alcohols lead to lignin. The enzyme is closely related to lignification, and chemical inhibition of its activity reduces the synthesis of lignins (Moesbacher et al., 1990; Mauch-Mani and Slusarenko, 1996). [1]
We find the sequence of CAD from Ocimum basilicum. Taxonomy of the organism follows as: [2]
Eukaryota, Viridiplantae, Streptophyta, Embryophyta, Tracheophyta, Spermatophyta, Magnoliophyta, eudicotyledons, Gunneridae, Pentapetalae, asterids, lamiids, Lamiales, Lamiaceae, Nepetoideae, Ocimeae, Ocimum.
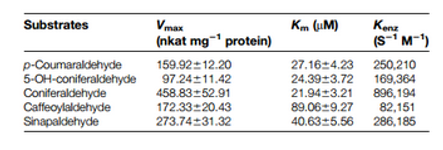
Figure 1.
Chemical Reaction of CAD:
Cinnamyl alcohol + NADP+ = cinnamaldehyde + NADPH.
Cofactor :Zn2+
Note: Binds 2 Zn2+ ions per subunit.
Functional expression of the Ocimum CAD1in E.coli PCR was used to introduce EcoRI , Xbal, Spel, Notl and Pstl at side ends of CAD1 coding region. The amplified product was single and double digested with EcoRl and Notl and the resultant fragment was cloned into the same sides ofthe puC57 vector. The engineered plasmid was transformed into E.coli BL 2l (DE3) cells which has lack of nudeases and proteinoses and ideal for expression. Proteins were extracted and then subjected to denaturating SDS-PAGE Tris-glycine gels and obtained protein gel results.
DNA gel analysis of CAD1 enzyme coding sequence. Each lone was loaded with 35 microliter DNA and the gel image obtained after usage of EcoRI and Not I restriction.

The predicted three-dimensional structure of CAD1. Data were analysed by the SWISS-MODEL software, and the figure was prepared with the program MOLMOL. a-helices are indicated in red and yellow, b-sheets by arrows, and turns and coils by curled lines. The NADPH binding and substrate binding sites are also indicated.
Figure 2. CAD1 Enzyme Part Resut
K1658006

Figure 3. 3D CAD1 Structure

Circular DNA form of CAD1+B0010(terminator) was obtained after plasmid isolation, here we can see them in last three lane. Those lanes were blurred but we have obtained more dense after reducing incubation time and increasing the amount of cell culture that we have used in plasmid isolation.
Figure 4. K1658002 Gel Result
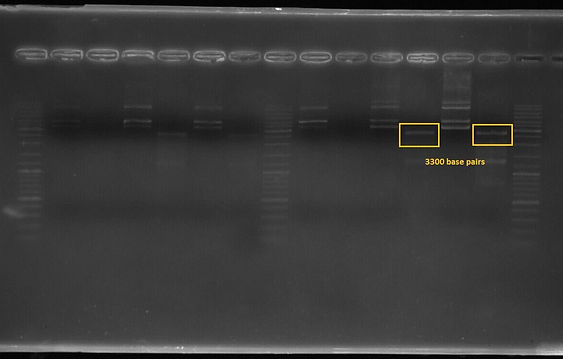
The first and third lanes before last ladder well shows that the composite part which includes constitutive promoter(j23100), Ribosome binding side(B0034), CAD1 enzyme coding gene and Terminator(B0010) in order at 3300 basepairs after double digestion.
Figure 5. K1658000 Gel Result
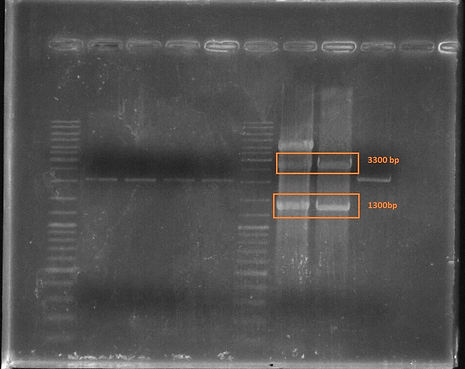
Here, we have obtained the double digested CAD1 enzyme coding gene with RBS and Terminator at approximately 1300 basepairs and after cloning on pSB1C3, double digestion gave us lanes at approximatle 3300 base pairs.
Figure 6. K1658003 Gel Result

As characterization of previously submitted parts, we have ligated RBS+Limonene synthase gene coding part and GFP reporter gene and obtain lane at approximatly 2500 basepairs after double digestion.
Figure 7. Limonane synthase K1658005 Gel Result

After ligation of I742111 and E0240, we have complete the circuit with constitutive promoter(J23116) and the difference between I742111+E0240 and J231116+I742111+E0240 can be seen by analyzing three lanes after ladder.
Figure 8. K1658004 Gel Result
Characterization of our part K1658000 by Fehling Experiment
 |  |
|---|---|
 |  |
 |
Fehling's solution is actually a mixture of two solution that are kept apart until needed. These are called Fehling's A and Fehling's B solutions.
Fehling's A is a solution of copper(II) sulphate and Fehling's B is a mixture of sodium hydroxide and potassium sodium tartrate (2,3-dihydroxybutanedioate).
When these two solutions are mixed together they form a copper(II) complex ion with the 2,3-dihydroxybutanedioate ligand. [Cu(tartrate)2]2+.
When the suspected aldehyde is warmed with a sample of this mixture, a red precipitate of copper(I) oxide is indicative of an aldehyde.
The copper(II) complex is reduced by the aldehyde to copper(I) oxide.
We used this technique to show that our CAD1 enzyme works and converts cinnamon alcohol to cinamaldehyde.
Since aldehyde is form, copper(II) complexes could be observed.

References
[1] Roth, R., Boudet, A., & Lezica, R. (1997, February 1). Lignification and cinnamyl alcohol dehydrogenase activity in developing stems of tomato and poplar: A spatial and kinetic study through tissue printing. Journal of Experimental Botany, 247-254.
[2] theeuropeanbioinformaticsinstitude/gene/np/http://www.ebi.ac.uk/
[3] Ma, Q. (2010). Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. Journal of Experimental Botany, 2735-2744. doi:10.1093/jxb/erq107
Figure 1. Ma, Q. (2010). Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. Journal of Experimental Botany, 2735-2744. doi:10.1093/jxb/erq107
Figure 3. Ma, Q. (2010). Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. Journal of Experimental Botany, 2735-2744. doi:10.1093/jxb/erq107